Tumors and centrosomes: Researchers receive grant to investigate cancer evolution

Unlike many of us during the COVID-19 pandemic, biological cells are not isolated from the outside world. Chemical variations, intercellular activity, and other microenvironmental factors impact cell survival. The relationship between cell and environment also applies to the development of cancer, which a team of Virginia Tech scientists is now researching.
After receiving a $1.4 million grant from the National Institutes of Health (NIH), researchers across multiple disciplines are launching a study to investigate tumor development within a natural environment.
With vital backing by the NIH, the study could provide information key to unravelling the mystery of cancer evolution and, in turn, establishing more effective cancer treatments.
“This research project focuses on the early evolution of cells after they double their genome. It is believed that many cancer cells go through this genome doubling during their evolution,” said Daniela Cimini, a professor from the Department of Biological Sciences in the College of Science. “We don’t know what happens in between the time the cancer cells double their genome and proliferation of a tumor.”
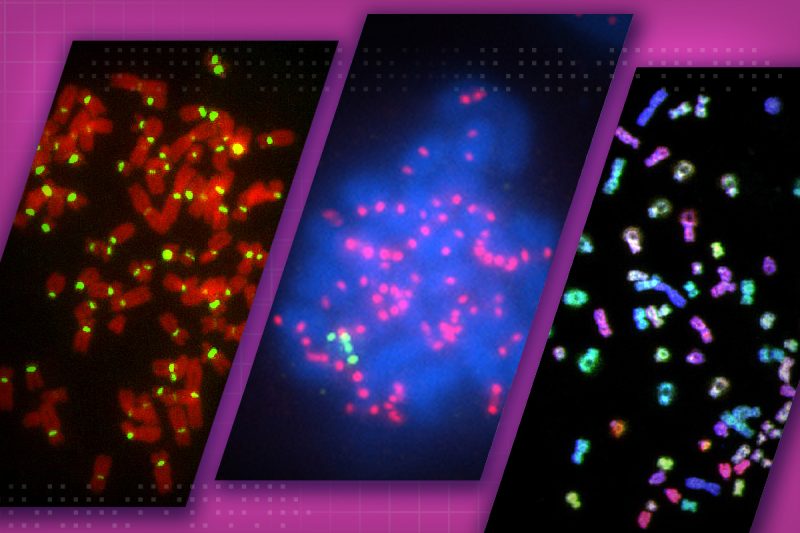
The unnatural genome doubling, called tetraploidization, is thought to be an intermediate step toward aneuploidy, or abnormal chromosome numbers. Instead of the usual 46 chromosomes, the cells feature at least one less or one too many chromosomes. Cancer cells almost always display the multiple chromosome characteristic.
Tetraploidy not only involves abnormal chromosome numbers but also doubles the cell’s centrosomes — subcellular complexes that are integral to cell division. These extra centrosomes may play a pivotal role in cancer development, but they were also found to disappear in standard cell culture conditions outside a living organism. The Cimini research group documented this in a prior study.
“In contrast to a petri dish, other factors within a living organism may change the extra centrosomes’ behavior to promote tumor development,” said Cimini, who is also co-director of the BIOTRANS program within the Fralin Life Sciences Institute.
Her team’s current research aims to solve the mystery of the disappearing centrosomes while studying the tumor microenvironment.
“Think of the tumor tissue as an ecosystem. The cancer cell really doesn’t exist by itself, but it exists in the context of this ecosystem,” said Scott Verbridge, a professor from the Department of Biomedical Engineering and Mechanics in the College of Engineering. “The development of malignant cancer is really characterized by tumor cells adapting to a very stressful tissue environment. The hope is that when you understand the dynamics of how the tumor cell becomes like it does in vivo, you then might be able to identify weak points or ways of targeting the cancer cell.”
By using mouse models and Verbridge’s tumor engineering, the project will focus on how chromosome and centrosome numbers change due to certain microenvironmental factors, such as oxygen levels within the tissue and the stiffness of the surface on which cells grow, known as substrate.
Crucial for the study, mathematical modeling will bolster the experimental processes and manage complex, dynamic data about the centrosomes and chromosomes.
“So, there are certain parts of the experiments that need to be done in vivo, and that’s very difficult,” said Jing Chen, a professor in the Department of Biological Sciences in the College of Science and an affiliated faculty member of the Fralin Life Sciences Institute. “If we have a model, we can use the model to simulate what’s going on and use a small amount of data to calibrate the model to make sure that it’s actually mimicking what’s happening in reality.”
Chen, an expert in mathematical modeling, worked with Cimini in the preceding study focused on tetraploid centrosomes. With an in vivo study, the mathematical modeling allows researchers to form new hypotheses that are testable in a natural setting.
“The model can give you the most likely reason for you seeing certain results,” Cimini said. “Then you can design an experiment to test if the reason suggested by the model is true. Instead of going in blind, the model helps prioritize which experiments are most likely to yield meaningful information and thereby, saves time and energy for experimentation.”
If scientists and medical professionals know what is important for tumor progression, they may know what weak points to target for future cancer treatments beyond just killing the cancer cells.
Traditional cancer therapies, although sometimes successful, involve toxic drugs that can also damage healthy cells and not affect the remaining, more lethal cancer cells.
“For the ovarian cancer cells, you kill some off and the others develop strategies to avoid toxicity,” said Eva Schmelz, a collaborator in the project, professor from the Department of Human Nutrition, Foods and Exercise in the College of Agriculture and Life Sciences and an affiliated faculty member of the Fralin Life Sciences Institute. “For one, that has a lot of side effects, but it’s also not that successful. Developing strategies to treat them earlier would be much better.”
With an emphasis on the microenvironment, the study could shed light on weak points that undermine cancer evolution before the cells even reach a malignant state.
Future research will be needed to test any potential therapies, but depending on its results, the project could reveal new targets for cancer treatment.
Receiving the NIH grant was made possible through several years of work by Cimini and her team to collect preliminary data and establish a framework. This was possible thanks to small seed grants, including those from the College of Science and the ICTAS Center for Engineered Health.
- Written by Tyler Harris
Related articles:




